Keywords: variance reduction | stratification | | Download Notebook
Contents
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
print("Finished Setup")
Finished Setup
The key idea in stratification is to split the domain on which we wish to calculate an expectation or integral into strata. Then, on each of these strata, we calculate the sub-integral as an expectation separately, using whatever method is appropriate for the stratum, and which gives us the lowest variance. These expectations are then combined together to get the final value.
In other words we can achieve better sampling in needed regions by going away from a one size fits all sampling scheme. One way to think about it is that regions with higher variability might need more samples, while not-so-varying regions could make do with less.
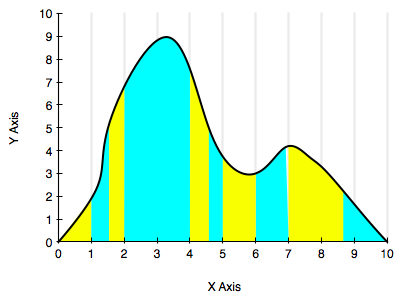
The diagram below illustrates this a bit. One could stratify by staying on the given grid, but the blue and yellow strata we have chosen might be better.
Now, instead of taking $N$ samples, we break the interval into $M$ strata and take $n_j$ samples for each strata $j$, such that $N=\sum_j n_j$.
Defining:
We can then define the stratified estimator of the overall expectation
which is an unbiased estimator of $\mu$.
where
is the “population variance” of $h(x)$ with respect to pdf $f_j(x)$ in region of support $D_j$.
Example
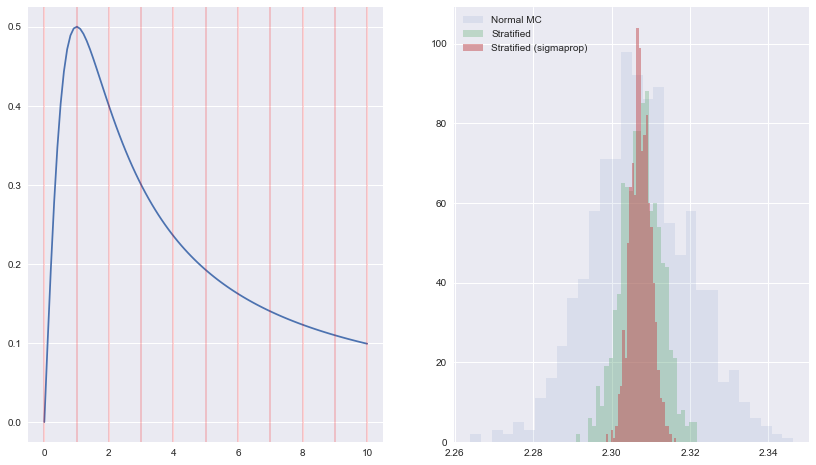
For a one-dimensional application we take $ x/(x^2+1)$ and integrate from $[0,1]$. We break $[0,10]$ into $M$ strata and for each stratum, take $N/M$ samples with uniform probability distribution. Compute the average within each stratum, and then calculate the overall average.
See http://www.public.iastate.edu/~mervyn/stat580/Notes/s09mc.pdf
plt.figure(figsize=[14,8])
Y = lambda x: x/(x**2+1.0);
intY = lambda x: np.log(x**2 + 1.0)/2.0;
N = 10000
Nrep = 1000
Ntry = 1000
Ns = 10 # number of strata
xmin=0
xmax =10
step = (xmax - xmin)/Ns
# analytic solution
Ic = intY(xmax)-intY(xmin)
Imc = np.zeros(Nrep)
Is = np.zeros(Nrep)
Is2 = np.zeros(Nrep)
## Ploting the original functions
plt.subplot(1,2,1)
x = np.linspace(0,10,100)
plt.plot(x, Y(x), label=u'$x/(x**2+1)$')
for j in range(Ns+1):
plt.axvline(xmin + j*step, 0, 1, color='r', alpha=0.2)
sigmas = np.zeros(Ns)
Utry = np.random.uniform(low=xmin, high=xmax, size=Ntry)
Ytry = Y(Utry)
Umin = 0
Umax = step
for reg in np.arange(0,Ns):
localmask = (Utry >= Umin) & (Utry < Umax)
sigmas[reg] = np.std(Ytry[localmask])
Umin = Umin + step
Umax = Umin + step
nums = np.ceil(N*sigmas/np.sum(sigmas)).astype(int)
print(sigmas, nums, np.sum(nums))
for k in np.arange(0,Nrep):
# First lets do it with mean MC method
U = np.random.uniform(low=xmin, high=xmax, size=N)
Imc[k] = (xmax-xmin)* np.mean(Y(U))
#stratified it in Ns regions
Umin = 0
Umax = step
Ii = 0
I2i = 0
for reg in np.arange(0,Ns):
x = np.random.uniform(low=Umin, high=Umax, size=N//Ns);
Ii = Ii + (Umax-Umin)*np.mean(Y(x))
x2 = np.random.uniform(low=Umin, high=Umax, size=nums[reg]);
I2i = I2i + (Umax-Umin)*np.mean(Y(x2))
Umin = Umin + step
Umax = Umin + step
Is[k] = Ii
Is2[k] = I2i
plt.subplot(1,2,2)
plt.hist(Imc,30, histtype='stepfilled', label=u'Normal MC', alpha=0.1)
plt.hist(Is, 30, histtype='stepfilled', label=u'Stratified', alpha=0.3)
plt.hist(Is2, 30, histtype='stepfilled', label=u'Stratified (sigmaprop)', alpha=0.5)
plt.legend()
print(np.std(Imc), np.std(Is), np.std(Is2))
[ 0.16531214 0.03284132 0.0292761 0.01743354 0.01195081 0.00838309
0.00645184 0.00485085 0.0036279 0.00303405] [5839 1160 1034 616 423 297 228 172 129 108] 10006
0.012575540594 0.00507214579121 0.002644516884