Keywords: regression | priors | uninformative priors | mcmc | bayesian | jeffreys prior | Download Notebook
Summary
Based on an example from the pymc3 docs and Jake Vanderplas's must-read blog article at http://jakevdp.github.io/blog/2014/03/11/frequentism-and-bayesianism-a-practical-intro/ , we do in pymc a custom regression with uninformative priors. We cover how to create custom densities and how to use theano shared variables to get posterior predictives.
Contents
%matplotlib inline
import numpy as np
import scipy as sp
import matplotlib as mpl
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import pandas as pd
pd.set_option('display.width', 500)
pd.set_option('display.max_columns', 100)
pd.set_option('display.notebook_repr_html', True)
import seaborn as sns
sns.set_style("whitegrid")
sns.set_context("poster")
Generate Data
This example is adapted from the pymc3 tutorial
np.random.seed(42)
theta_true = (25, 0.5)
xdata = 100 * np.random.random(20)
ydata = theta_true[0] + theta_true[1] * xdata
# add scatter to points
xdata = np.random.normal(xdata, 10)
ydata = np.random.normal(ydata, 10)
We’ll take a detour to be able to posterior predict elsewhere
from theano import shared
xdata_shared = shared(xdata)
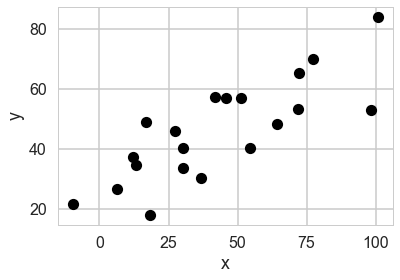
plt.plot(xdata, ydata, 'ok')
plt.xlabel('x')
plt.ylabel('y');
import pymc3 as pm
Jeffrey’s priors
What do we do for priors of scale variables, such as the precision/variance of a gaussian?
The point of Jeffreys Priors is to create priors that don’t become informative when you transform the variables. Jeffreys Priors are defined as:
The Fisher Information $I$ is the second derivative of the likelihood surface and thus represents the curvature of that surface:
where the expectation is with respect to the sampling distribution (the likelihood as a distribution).
You can convince yourself that $p_J$ is invariant under a change of variable, unlike the uniform prior; this is particularly easy to show in one dimension.
Jeffrey’s priors for the gaussian-gaussian model
For a gaussian with known $\sigma$ (see the normal model), lets find the Jeffrey’s prior for the location parameter $\mu$:
(by $f \mid \sigma$ we mean holding $\sigma$ fixed or likelihood as a function of $\mu$)
The Jeffrey’s prior for $\mu$ is thus $1/\sigma$ and for fixed $\sigma$ this is the improper uniform….
What about for $\sigma$:
Then:
which makes the prior $1/\sigma$.
Writing the model
Setting priors
Where do the priors for $\sigma$ and $\beta$ come from? The prior for $\sigma$, as we have seen earlier, is the Jeffrey’s prior on $\sigma$ the scale parameter for a normal-normal model.
The prior on $\beta$ the slope (remember, we dont want to put a uniform prior on slope) can be derived as an uninformative prior based on symmetry, say:
Thus $\alpha^{‘} = \beta/\alpha$, and $\beta^{‘} = 1/\beta$.
The jacobian of the transformation is $\beta^3$ so that $q(\alpha^{‘}, \beta^{‘}) = \beta^3 p(\alpha, \beta)$ where $q$ is the pdf in the transformed variables. But now we want uninformativeness, or that the transformation should not affect the pdf, and we can check that the prior we chose makes $q=p$ in functional form:
We will need to write custom densities for this. Theano provides us a way:
import theano.tensor as T
with pm.Model() as model1:
alpha = pm.Normal('intercept', 0, 100)
# Create custom densities, you must supply logp
beta = pm.DensityDist('beta', lambda value: -1.5 * T.log(1 + value**2), testval=10)
eps = pm.DensityDist('eps', lambda value: -T.log(T.abs_(value)), testval=10)
# Create likelihood
like = pm.Normal('y_est', mu=alpha + beta * (xdata_shared - xdata_shared.mean()), sd=eps, observed=ydata)
for RV in model1.basic_RVs:
print(RV.name, RV.logp(model1.test_point))
intercept -5.524108719192764
beta -6.92268077526189
eps -2.302585092994046
y_est -8385.484891686912
Sampling
And now we sample..
with model1:
stepper=pm.Metropolis()
tracem1 = pm.sample(100000, step=stepper, njobs=2)
#tracem1 = pm.sample(4000)
Multiprocess sampling (2 chains in 2 jobs)
CompoundStep
>Metropolis: [eps]
>Metropolis: [beta]
>Metropolis: [intercept]
Could not pickle model, sampling singlethreaded.
Sequential sampling (2 chains in 1 job)
CompoundStep
>Metropolis: [eps]
>Metropolis: [beta]
>Metropolis: [intercept]
100%|██████████| 100500/100500 [00:49<00:00, 2011.49it/s]
100%|██████████| 100500/100500 [00:51<00:00, 1933.72it/s]
The number of effective samples is smaller than 25% for some parameters.
tm1=tracem1[40000::]
pm.traceplot(tm1);
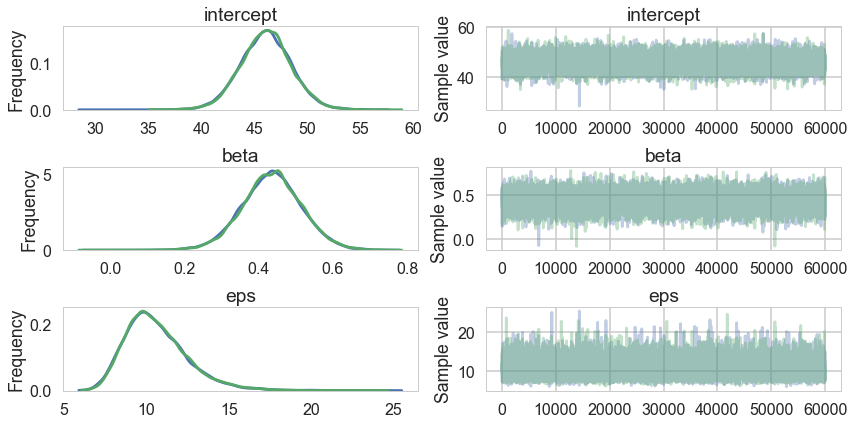
pm.summary(tm1)
| mean | sd | mc_error | hpd_2.5 | hpd_97.5 | n_eff | Rhat | |
|---|---|---|---|---|---|---|---|
| intercept | 46.082865 | 2.412582 | 0.013754 | 41.331279 | 50.789888 | 25957.0 | 1.000042 |
| beta | 0.433899 | 0.081065 | 0.000531 | 0.274618 | 0.595071 | 25344.0 | 0.999998 |
| eps | 10.634574 | 1.915740 | 0.015013 | 7.312843 | 14.458126 | 17637.0 | 1.000025 |
pm.autocorrplot(tm1);
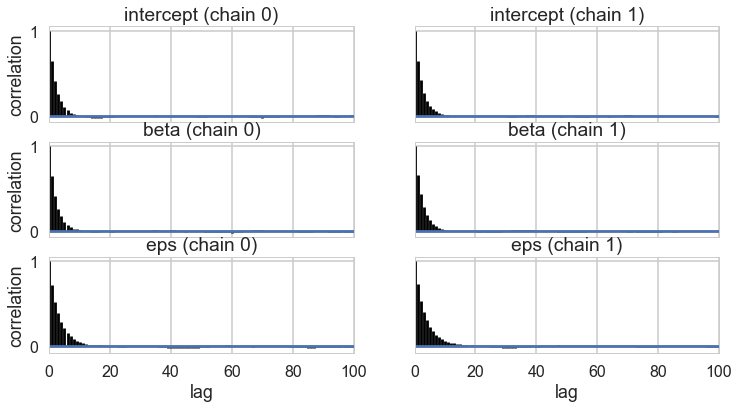
pm.trace_to_dataframe(tm1).corr()
| intercept | beta | eps | |
|---|---|---|---|
| intercept | 1.000000 | 0.001150 | 0.004989 |
| beta | 0.001150 | 1.000000 | -0.042344 |
| eps | 0.004989 | -0.042344 | 1.000000 |
Results
def plot_MCMC_model(ax, xdata, ydata, trace):
"""Plot the linear model"""
ax.plot(xdata, ydata, 'ok')
intercept, beta = trace['intercept'][:,None], trace['beta'][:,None]
xfit = np.linspace(-20, 120, 10)
yfit = intercept + beta * (xfit - xfit.mean())
mu = yfit.mean(0)
sig = 2 * yfit.std(0)
ax.plot(xfit, mu, '-k')
ax.fill_between(xfit, mu - sig, mu + sig, color='lightgray')
ax.set_xlabel('x')
ax.set_ylabel('y')
def compute_sigma_level(trace1, trace2, nbins=20):
"""From a set of traces, bin by number of standard deviations"""
L, xbins, ybins = np.histogram2d(trace1, trace2, nbins)
L[L == 0] = 1E-16
logL = np.log(L)
shape = L.shape
L = L.ravel()
# obtain the indices to sort and unsort the flattened array
i_sort = np.argsort(L)[::-1]
i_unsort = np.argsort(i_sort)
L_cumsum = L[i_sort].cumsum()
L_cumsum /= L_cumsum[-1]
xbins = 0.5 * (xbins[1:] + xbins[:-1])
ybins = 0.5 * (ybins[1:] + ybins[:-1])
return xbins, ybins, L_cumsum[i_unsort].reshape(shape)
def plot_MCMC_trace(ax, xdata, ydata, trace, scatter=False, **kwargs):
"""Plot traces and contours"""
xbins, ybins, sigma = compute_sigma_level(trace['intercept'], trace['beta'])
ax.contour(xbins, ybins, sigma.T, levels=[0.683, 0.955], **kwargs)
if scatter:
ax.plot(trace['intercept'], trace['beta'], ',k', alpha=0.1)
ax.set_xlabel(r'intercept')
ax.set_ylabel(r'$\beta$')
def plot_MCMC_results(xdata, ydata, trace, colors='k'):
"""Plot both the trace and the model together"""
fig, ax = plt.subplots(1, 2, figsize=(10, 4))
plot_MCMC_trace(ax[0], xdata, ydata, trace, True, colors=colors)
plot_MCMC_model(ax[1], xdata, ydata, trace)
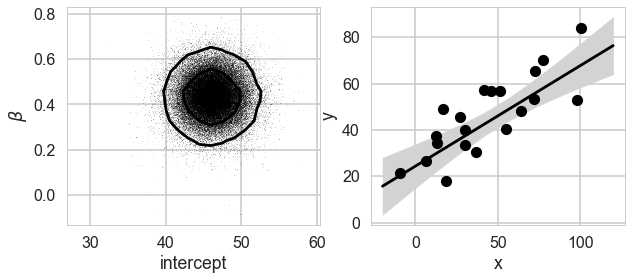
We can plot our posterior contours and see how the posterior reacts to our data.
plot_MCMC_results(xdata, ydata, tm1)
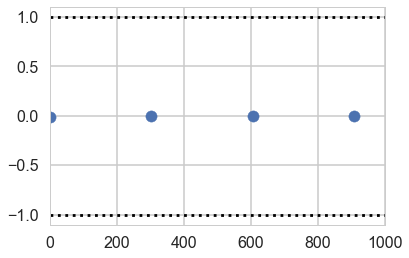
Gewecke convergence test
Our sample-runs give use per-run z scores for the difference of means wau=y within the 1 to -1 bracket.
z = pm.geweke(tm1, intervals=100);
plt.scatter(*z[0]['intercept'].T)
plt.hlines([-1,1], 0, 1000, linestyles='dotted')
plt.xlim(0, 1000)
(0, 1000)
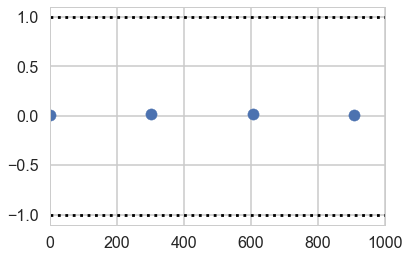
plt.scatter(*z[0]['beta'].T)
plt.hlines([-1,1], 0, 1000, linestyles='dotted')
plt.xlim(0, 1000)
(0, 1000)
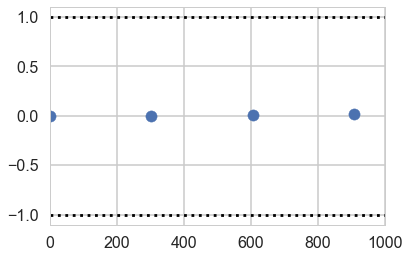
plt.scatter(*z[1]['eps'].T)
plt.hlines([-1,1], 0, 1000, linestyles='dotted')
plt.xlim(0, 1000)
(0, 1000)
Getting the posterior predictive
xdata_oos=np.arange(-20, 120,1) #out of sample
xdata_shared.set_value(xdata_oos)
ppc = pm.sample_ppc(tm1, model=model1, samples=500)
100%|██████████| 500/500 [00:03<00:00, 146.72it/s]
ppc['y_est'].shape, xdata.shape, xdata_oos.shape
((500, 140), (20,), (140,))
yppc = ppc['y_est'].mean(axis=0)
yppcstd=ppc['y_est'].std(axis=0)
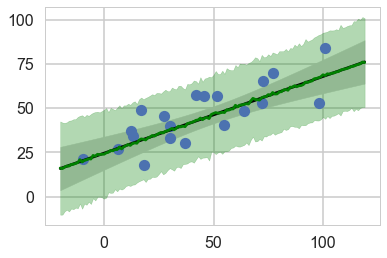
plt.plot(xdata, ydata,'o');
intercept, beta = tm1['intercept'][:,None], tm1['beta'][:,None]
yfit = intercept + beta * (xdata_oos - xdata_oos.mean())
mu = yfit.mean(0)
sig = 2 * yfit.std(0)
plt.plot(xdata_oos, mu, '-k')
plt.fill_between(xdata_oos, mu - sig, mu + sig, color='lightgray')
plt.plot(xdata_oos, yppc, color="green")
plt.fill_between(xdata_oos, yppc - 2*yppcstd, yppc + 2*yppcstd, color='green', alpha=0.3)
<matplotlib.collections.PolyCollection at 0x11309eb00>