Keywords: identifiability | mcmc | bayesian | Download Notebook
Contents
Contents
%matplotlib inline
import numpy as np
import scipy as sp
import matplotlib as mpl
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import pandas as pd
pd.set_option('display.width', 500)
pd.set_option('display.max_columns', 100)
pd.set_option('display.notebook_repr_html', True)
import seaborn as sns
sns.set_style("whitegrid")
sns.set_context("poster")
We generate some test data from $N(0,1)$:
from scipy.stats import norm
data = norm.rvs(size=100)
data
array([ 0.03962073, 0.03290953, 0.98544195, 0.04596463, 0.64626776,
-1.51917579, -0.47099774, -0.79274817, -1.510941 , 0.62187968,
-1.58463447, -0.31167464, 1.09177345, 1.86007596, -0.34114228,
-0.35475686, 0.70235072, -0.87451605, -1.14164176, -1.33450384,
-1.66049335, 0.42916234, -1.45189161, -1.80733019, -1.34731763,
0.5692819 , -1.54416497, 0.06534625, -1.42065169, -0.20115099,
1.13634278, 0.33987116, 0.40928055, -0.79304293, -1.42792586,
-0.49092787, -0.40731762, 0.36158092, 0.55836612, 0.94629738,
0.37472964, 0.67798211, 0.47590639, -0.00241526, 1.84591318,
0.24104702, 0.22500321, -1.71406405, 2.17130125, -0.4478053 ,
0.51994149, -0.56184292, 0.2997452 , -0.27366469, -0.90988938,
1.0146704 , -0.99058279, -0.29684026, -1.43256059, 0.03973359,
1.09712035, 0.46719219, 0.80223718, 1.54859038, -0.94135931,
-0.20712364, -0.21525818, -0.84091686, 0.16319026, -0.35566993,
-0.17525355, -0.13103397, -0.9963371 , 0.62021913, -0.29783109,
-1.4712209 , -0.06468791, 0.85275671, 1.48148843, 0.15819005,
1.45768006, -0.536146 , -0.01650962, -0.07422125, -0.33416329,
1.64273025, 0.55148576, 1.14837232, -0.37978736, -0.57564425,
-0.11643753, 0.52559572, -1.73441951, -0.79981563, 0.0109395 ,
-0.93453305, -0.46059307, -2.31094356, -1.03155391, 0.22409766])
We fit this data using the following model:
import pymc3 as pm
In our sampler, we have chosen njobs=2 which allows us to run on multiple processes, generating two separate chains.
with pm.Model() as ni:
sigma = pm.HalfCauchy("sigma", beta=1)
alpha1=pm.Uniform('alpha1', lower=-10**6, upper=10**6)
alpha2=pm.Uniform('alpha2', lower=-10**6, upper=10**6)
mu = pm.Deterministic("mu", alpha1 + alpha2)
y = pm.Normal("data", mu=mu, sd=sigma, observed=data)
stepper=pm.Metropolis()
traceni = pm.sample(100000, step=stepper, njobs=2)
100%|██████████| 100000/100000 [01:17<00:00, 1286.59it/s]| 1/100000 [00:00<3:20:31, 8.31it/s]
pm.traceplot(traceni);
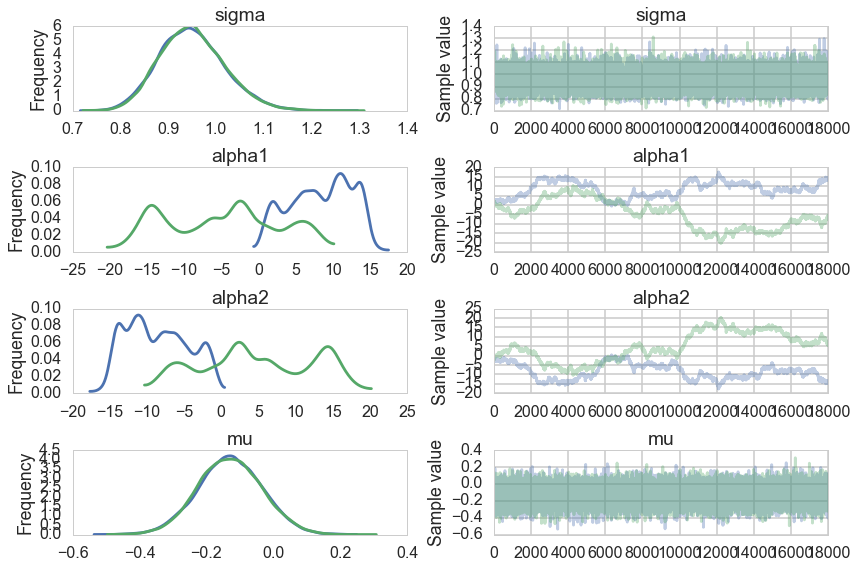
Look at our traces for $\alpha_1$ and $\alpha_2$. These are bad, and worse, they look entirely different for two chains. Despite this, $\mu$ looks totally fine. Our trac
df=pm.trace_to_dataframe(traceni)
df.corr()
| sigma | mu | alpha1 | alpha2 | |
|---|---|---|---|---|
| sigma | 1.000000 | -0.000115 | -0.003153 | 0.003152 |
| mu | -0.000115 | 1.000000 | 0.002844 | 0.008293 |
| alpha1 | -0.003153 | 0.002844 | 1.000000 | -0.999938 |
| alpha2 | 0.003152 | 0.008293 | -0.999938 | 1.000000 |
Just like in our uncentered regression example, we have $\alpha_1$ and $\alpha_2$ sharing information: they are totally negatively correlated and unidentifiable. Indeed our intuition probably told us as much.
pm.summary(traceni)
sigma:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.950 0.068 0.000 [0.821, 1.086]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.827 0.903 0.947 0.994 1.095
alpha1:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
1.730 9.037 0.897 [-15.433, 14.707]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
-16.135 -4.079 3.764 8.813 14.291
alpha2:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
-1.859 9.038 0.897 [-14.814, 15.325]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
-14.420 -8.950 -3.904 3.960 16.012
mu:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
-0.130 0.096 0.001 [-0.319, 0.057]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
-0.317 -0.195 -0.130 -0.065 0.059
A look at the effective number of samples using two chains tells us that we have only one effective sample for $\alpha_1$ and $\alpha_2$.
pm.effective_n(traceni)
{'alpha1': 1.0,
'alpha1_interval_': 1.0,
'alpha2': 1.0,
'alpha2_interval_': 1.0,
'mu': 26411.0,
'sigma': 39215.0,
'sigma_log_': 39301.0}
The Gelman-Rubin statistic is awful for them. No convergence.
pm.gelman_rubin(traceni)
{'alpha1': 1.7439881580327452,
'alpha1_interval_': 1.7439881580160093,
'alpha2': 1.7438626593529831,
'alpha2_interval_': 1.7438626593368223,
'mu': 0.99999710182062695,
'sigma': 1.0000248056117549,
'sigma_log_': 1.0000261752214563}
Its going to be hard to break this unidentifiability. We try by forcing $\alpha_2$ to be negative in our prior
with pm.Model() as ni2:
sigma = pm.HalfCauchy("sigma", beta=1)
alpha1=pm.Normal('alpha1', mu=5, sd=1)
alpha2=pm.Normal('alpha2', mu=-5, sd=1)
mu = pm.Deterministic("mu", alpha1 + alpha2)
y = pm.Normal("data", mu=mu, sd=sigma, observed=data)
#stepper=pm.Metropolis()
#traceni2 = pm.sample(100000, step=stepper, njobs=2)
traceni2 = pm.sample(100000, njobs=2)
Average ELBO = -143.13: 100%|██████████| 200000/200000 [00:18<00:00, 10759.64it/s], 9912.87it/s]
100%|██████████| 100000/100000 [06:30<00:00, 255.83it/s]
Notice we are using the built in NUTS sampler. It takes longer but explores the distributions far better: more on this after our course break.
pm.traceplot(traceni2);
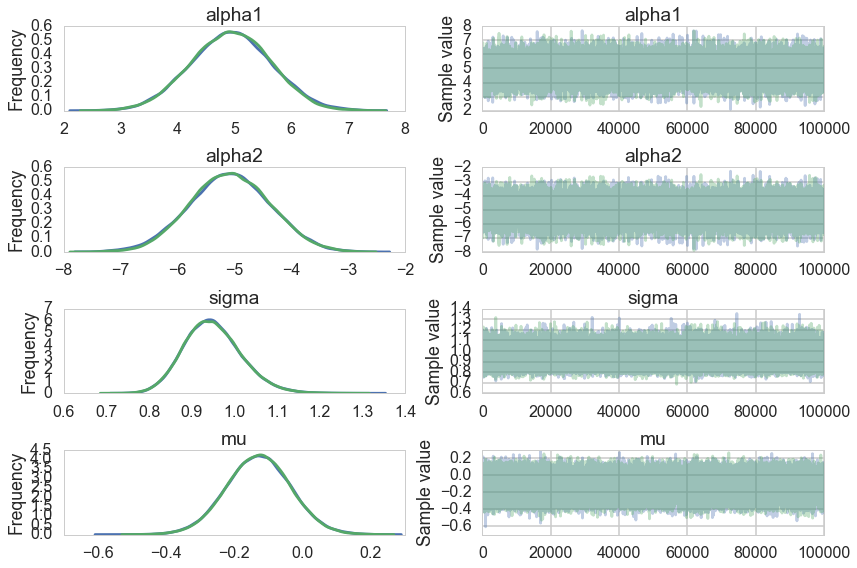
Our extremely strong priors have helped us do a much better job.
pm.summary(traceni2)
alpha1:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
4.944 0.707 0.004 [3.560, 6.314]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
3.562 4.465 4.946 5.424 6.319
alpha2:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
-5.074 0.707 0.004 [-6.471, -3.712]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
-6.450 -5.552 -5.076 -4.593 -3.690
sigma:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
0.950 0.067 0.000 [0.820, 1.083]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
0.828 0.903 0.946 0.992 1.093
mu:
Mean SD MC Error 95% HPD interval
-------------------------------------------------------------------
-0.129 0.094 0.000 [-0.313, 0.057]
Posterior quantiles:
2.5 25 50 75 97.5
|--------------|==============|==============|--------------|
-0.315 -0.192 -0.129 -0.066 0.056
Our effective sample size is still poor and our traces still look dodgy, but things are better.
pm.effective_n(traceni2)
{'alpha1': 21779.0,
'alpha2': 21771.0,
'mu': 200000.0,
'sigma': 155001.0,
'sigma_log_': 156987.0}
pm.gelman_rubin(traceni2)
{'alpha1': 1.0000646992489557,
'alpha2': 1.0000681135686484,
'mu': 0.99999730757810168,
'sigma': 1.0000011955172152,
'sigma_log_': 1.000002093954572}
..and this shows in our Gelman-Rubin statistics as well…
pm.trace_to_dataframe(traceni2).corr()
| alpha1 | alpha2 | mu | sigma | |
|---|---|---|---|---|
| alpha1 | 1.000000 | -0.991091 | 0.066087 | 0.000749 |
| alpha2 | -0.991091 | 1.000000 | 0.067395 | -0.000322 |
| mu | 0.066087 | 0.067395 | 1.000000 | 0.003204 |
| sigma | 0.000749 | -0.000322 | 0.003204 | 1.000000 |
..but our unidentifiability is still high when we look at the correlation. This reflects the fundamental un-identifiability and sharing of information in our model since $\mu = \alpha_1 +\alpha_2$: all the priors do is artificially peg one of the parameters. And once one is pegged the other is too because of the symmetry.